5 TYPES OF INTERMOLECULAR INTERACTIONS
Beside the protein-DNA interactions discussed previously , 5 protein-protein interaction surfaces exist within the tetramers, and contribute to the p73 monomer-dimer-tetramer assembly route (see Figure 9)
A)Two monomer-monomer interaction surfaces, A-B and C-D, stabilize the dimer.
B) Three dimer-dimer interactions surfaces, A-D, B-D and B-D, form the tetramer.
 |
| Figure 9: Different types of interactions within the tetramer |
DIMERIZATION INTERFACES
Dimerization interfaces are not affected by spacer insertion. Two distinct dimer conformations exist: one influenced by tetramerization (in the cases of 0 and 1bp spacers) and one independent from it (in the cases of 2 and 4bp spacers, where tetramerization restraints are weak or absent).
The dimerization interface seems to be able to establish two different H-bond networks to produce these two conformations.
TETRAMERIZATION INTERFACES
Hydrogen bond contacts and hydrophobic interactions from residues located in monomer loops are the main actors of tetramerization interfaces.
For every base pair added in the spacer sequence, the distance between two dimers increases by 3.4Å (see Figure 10). For 0 and 1bp spacers, tetramerization interfaces can still affect the dimers. However, in the case of 2 and 4bp spacers, where the dimers are separated by 40 and 52Å respectively, the number of hydrogen bonds and hydrophobic contacts decreases, causing weak or no tetramerization interaction.
Different rotation angles between dimers cause the 0 and 1bp spacer tetramers to keep a flat dimer-to-dimer structure whereas the 2bp tetramer is not flat, due to the dimers rotating out of plane.
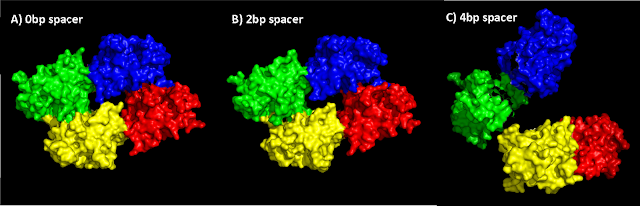 |
| Figure 10: Different tetramer conformations depending on spacer length. The longer the spacer sequence, the further apart dimers move. |
DNA CONFORMATIONS
The DNA structure of the spacer regions varies according to spacer length (see Figure 11).These DNA structural variations further influence protein-protein interactions and affect the overall conformation of the tetramer.
- For 0, the DNA adopts a classical B DNA (right handed, 10 base pairs per turn)
- 1 bp spacer DNA structures include an unwinding of the DNA double helix in the middle of the spacer and undergo a -30° twist at the center of the spacer. Moreover, a slight bending towards the major groove of the DNA allows the presence of an extra base without distorting the tetramer.
- 2bp spacer DNA structures include an unwinding of the DNA double helix too. The DNA molecule bends slightly but not enough to accommodate two extra bases without distorting the tetramer. The double helix unwinding is the key element allowing the tetramer to form despite the additional base pairs.
- 4bp spacer adopts a general B DNA structure but has a 3Å slide in the middle of the spacer.

No comments:
Post a Comment